SAPIENS – A New Generation MR Spectroscopy Processing, Analysis and Visualization Software
The Honours Project was done under the joint supervision of Arash Bakhtiari (CSE), Ralph Noeske, Timo Schirmer and Rolf Schulte (General Electric)
Magnetic Resonance Spectroscopy (MRS) is an important tool in modern day medical research. To process and interpret data which is obtained during such a MRS scan, computer software is necessary. Existing software solutions, however, may lack usability and modern design standards. The Project was conducted in cooperation with the General Electric Company (GE). Their MRS software Spectroscopy Analysis of GE (SAGE) required improvement in terms of usability, as well as from a software engineering point of view. The main objective of the project was to design a C/C++ based user-friendly MRS software with similar functionalities as SAGE.
Motivation
MRS is a non-invasive technology to visualize the concentration of metabolites in the tissue at specific locations in the human body. Similar to Magnetic Resonance Imaging (MRI) the method aquires information from the physical phenomenon of Nuclear Magnetic Resonance. While MRI measures mainly the free induction decay (FID) of hydrogen in water and fat, MRS is used to measure the FID of several metabolites for a given voxel. As a consequence, MRS allows to investigate different diseases like cancer, epilepsy, Alzheimer’s disease and Parkinson’s disease based on the anomalies of concentration of the metabolites in these voxels.
In comparison to MRI, which is a widely-used tool in clinical practise, the applicability of MRS is mainly limited to research institutions. One reason for this is its complexity in data acquisition, processing, analysis and visualization. The Honours Project tackles this problem by providing a highly extensible, maintainable, robust and pattern-based software called Sapiens. The software implements the functionality of the existing software SAGE in a contemporary way of programming using C++.
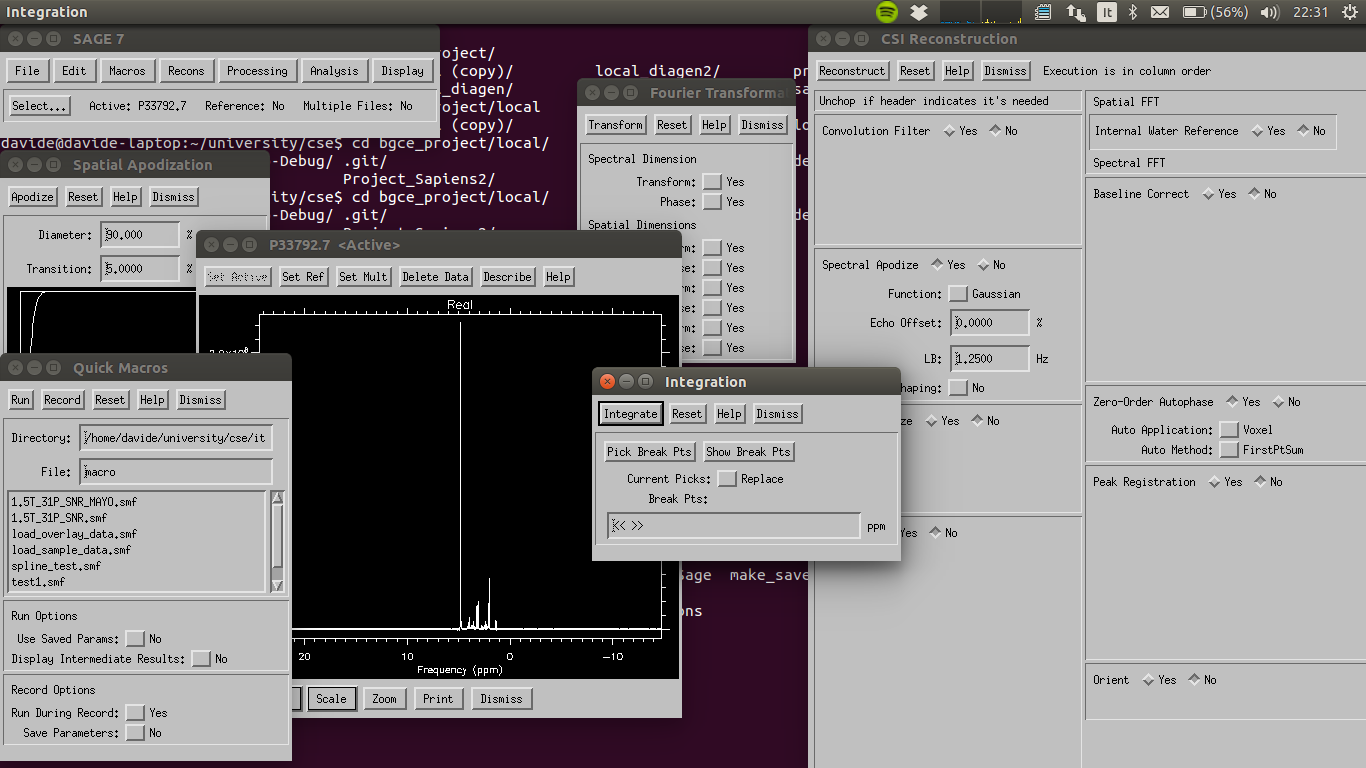
Figure 1: A normal usage situation of the SAGE software, showing the multiple opened secondary windows and the obsolete design
Development overview
The original software lacked appropriate documentation and usability. Therefore, a working infrastructure (specifically, Trac and GIT) to act as both ongoing, and final framework for progress and documentation of the new software was set up. The first main task of the project was to create the software design. This was done using detailed hierarchical UML diagrams with known software engineering patterns. Based on this, the core of the program was written in parallel with the development of the new Graphical User Interface (GUI). The solution was modular, meaning that after this core was done, program features required by GE could be easily added with little extra code.
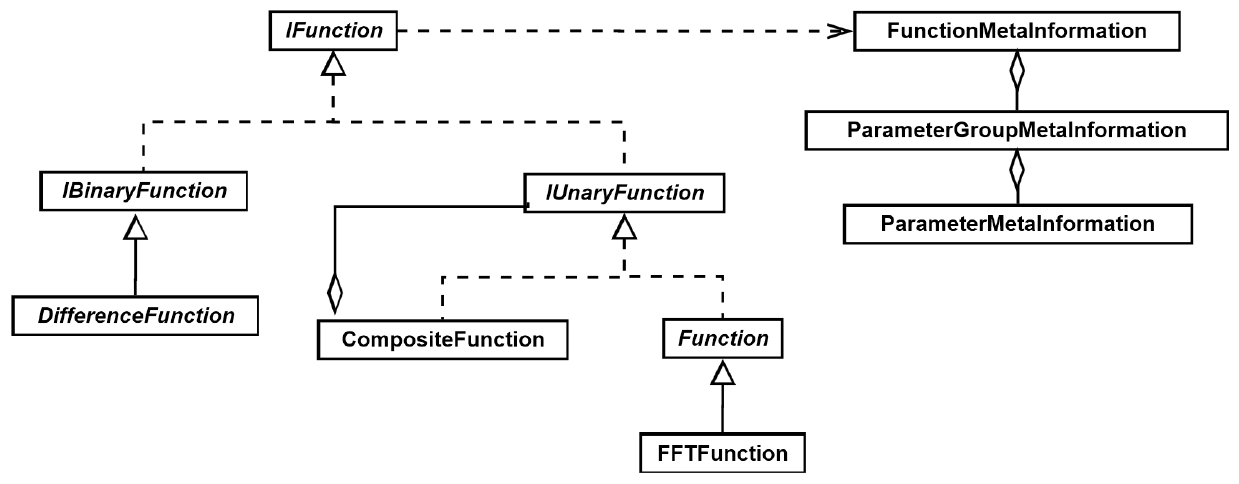
Figure 2: An example of the UML diagram of a single module (class), as used in the software design development
Features of the final software
- Data I/O operations on legacy SAGE files and DICOM format
- Computation: FFT, dataset substractions & additions
- Functions: Macros recording, undo/redo capabilities
- Interfacing: execution of MATLAB code
- Extensive code documentation
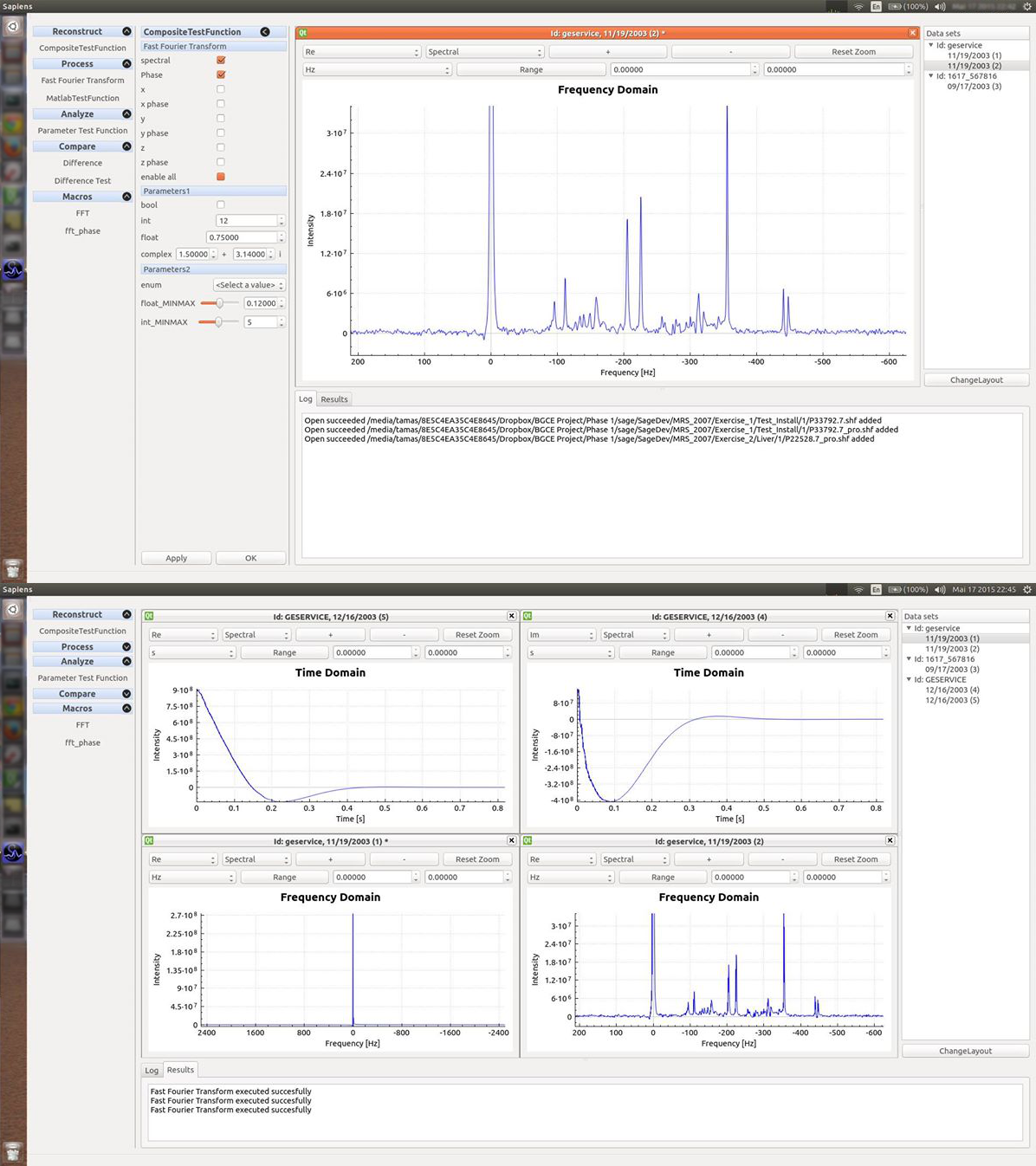
Figure 3: Two views of the new Sapiens software in usage, showing the modern look, intuitive design, and useful features
