under supervision by
Dr. Daniel Butnaru (Carl Zeiss Microscopy)
M.Sc. with honors Arash Bakhtiari (TUM)
Motivation
Modern microscopes rapidly generate massive quantities of raw image data. The conversion of this raw data into images which a human can interpret is a major task for the microscope system. In current setups, each microscope is connected to a single computer, which performs both acquisition and post processing of the acquired images. These tasks are run sequentially, which means that the computer cannot start post processing until all acquisition is finished, incurring an unnecessary delay in between setting experiment parameters and viewing the results. This time delay artificially restricts the user’s workflow, impeding interactivity and discouraging the user from collecting any more data than is strictly necessary.
This project aims to address the issues mentioned above by distributing the post processing over a cluster. A key goal is to provide the microscope user with tools for utilizing the cluster without requiring any knowledge of distributed systems. The network location, configuration, and inner workings of the cluster are kept transparent to the user. Instead, the user interacts with a domain-specific language which emphasizes concepts specific to the Zeiss image processing libraries and acquisition workflow. The user may chain together different Zeiss library functions, submit this ‘topology’ to the abstract cluster, send it data to process in real-time, and track its progress and performance.
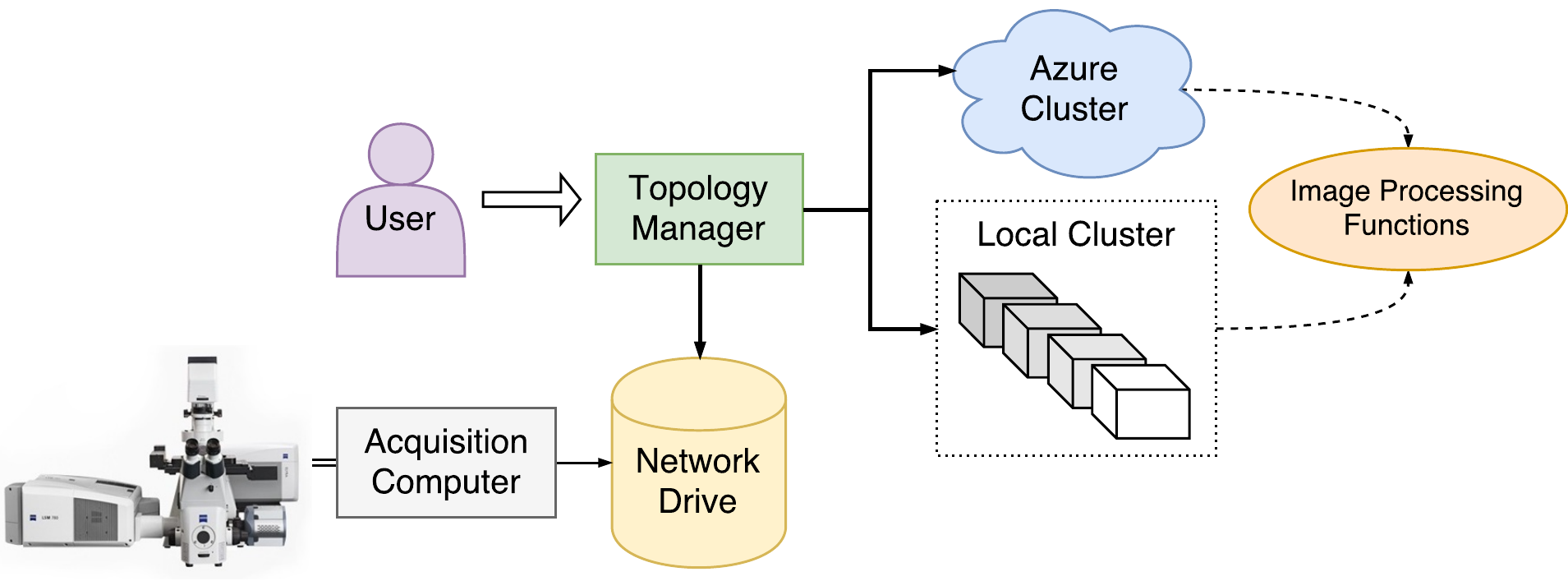
Behind the scenes, a server translates the user’s requests into interactions with an Apache Storm cluster. This framework was chosen because it leverages existing technologies and cloud infrastructure, simplifying management and maintenance. As a consequence, the cluster may be built on-site using older PCs, may be provided by Zeiss as a service, or may be automatically provisioned via AWS or Microsoft Azure. A high-level view of the system is shown in Figure 1.
Architecture
The project architecture consists of three subsystems which were developed in parallel: a frontend library which runs on each user’s computer, a middleware “TopologyManager” which serves an API from the edge node of the cluster, and a backend which wraps the Zeiss image processing libraries in the idioms of the Storm framework. The frontend is a CSharp DLL designed to be called from an interactive IronPython session or integrated directly into Zeiss’s microscope software. The TopologyManager, a Java ZeroMQ server, acts as a gateway to the cluster, allowing multiple clients to submit and control multiple topologies, notify a topology of new data, and view status, metrics, and event information. In order to run a topology, Storm instantiates, configures, and deploys different backend components as specified by the user in a topology definition. A builder interface was designed to make it intuitive for the user to define different topologies without extensive knowledge of the backend components. Meanwhile, the backend components were designed to run Zeiss libraries as generically as possible, including the option to submit user-written IronPython scripts which call the Zeiss libraries arbitrarily.
Topology Examples
The system directly supports Zeiss’s image segmentation routines. This is demonstrated by the CellCounter topology. CellCounter takes as input a file containing a 2D grid of images. Each image depicts one well of a well plate, in which some number of cells are growing. The system segments each image, counts the number of cells it detected in each well, and generates a heatmap of cell counts over the entire wellplate. This is depicted in Figure 2.
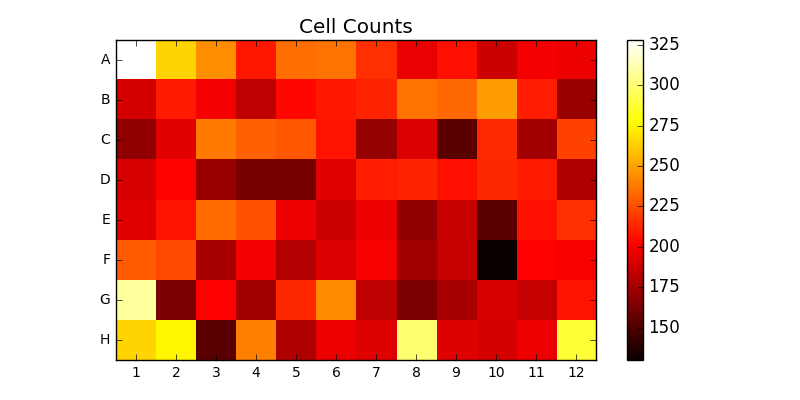
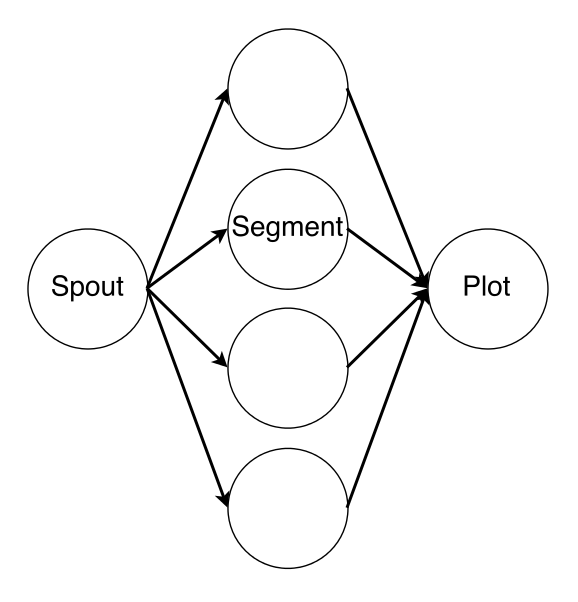
CellCounter is implemented using three different components: a PushSpout waits for notifications that new files are ready, before distributing them to the next available AnalyzerBolt, which performs the segmentation, before passing the results a custom ScriptBolt which generates the heatmap via Python and Matplotlib. Because the computational effort of the image segmentation is considerably higher than that of the plot generation, the majority of the parallelism is assigned to the AnalyzerBolt. The connectivity of the deployed topology is depicted in Figure 3.
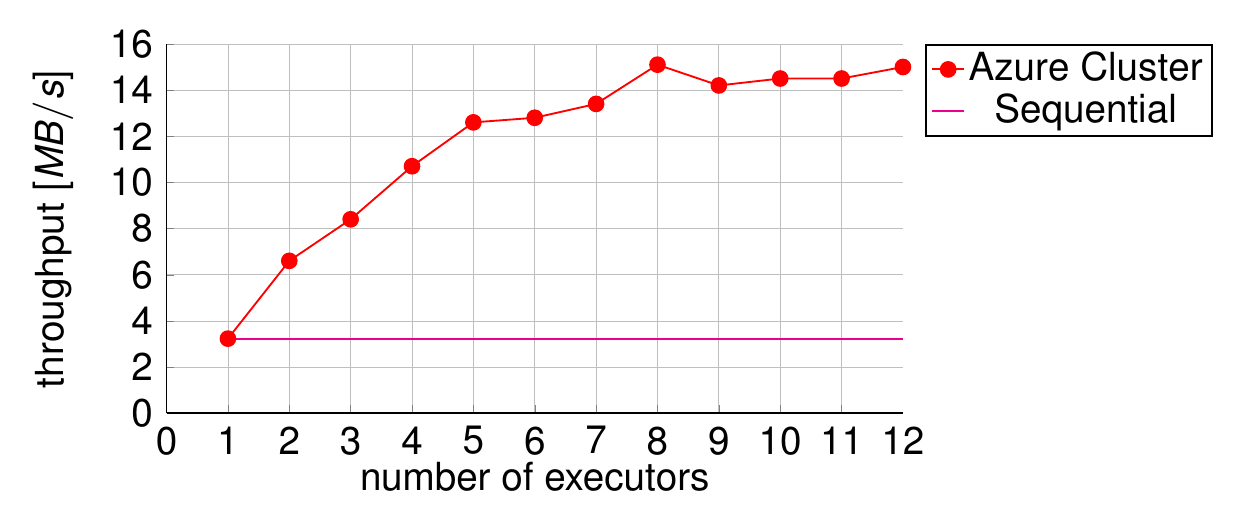
This topology was used to test the overall performance of the system. A stock Storm cluster was provisioned on Microsoft Azure and scaled to 4 worker machines. A test harness was built to feed the cluster as much data as it could handle and measure its steady-state performance. Compared to the sequential version, the overhead of messaging and file transfer added a latency of ~30 seconds, and this number remained stable as more worker threads were added. Meanwhile, the throughput improved from ~4 MB/s to ~15 MB/s, demonstrating that a much greater volume of data can be processed in the same amount of time. This is shown in Figure 4.